How drastically
different families of dUTPases interact with the same inhibitor protein: structural
insights
Kinga Nyíri1,2, Judit Matejka1,2, Matthew J. Harris3, Antoni J. Borysik3, Beáta G. Vértessy1,2
1Department of Applied Biotechnology and Food Science, Budapest Univ. of Technology and Economics, 4 Szent Gellért tér, Budapest, Hungary, 1111
2Institute of Enzymology, RCNS, Hungarian Academy of Sciences, 2 Magyar tudósok körútja, Budapest, Hungary, 1117
3Department of Chemistry, King’s College London, Britannia House, London SE1 1DB, United Kingdom
nyiri.kinga@ttk.mta.hu
The genetic material encoding all the machinery of a living organism is prone to several sorts of chemical modifications. Removal of the unsought modifications formed by spontaneous processes or induced by damaging agents is of paramount importance. The presence of uracil is one of the common errors, therefore several mechanisms exist to remove this base from DNA. As a part of preventive repair, dUTPase protein encumbers uracil misincorporation into DNA by hydrolysis of dUTP. This important role of dUTPase entitles it as an essential enzyme in most organisms, and also as a target of antiparasite drugs and cancer therapy [1].
It has been shown that Stl, a Staphylococcus
aureus pathogenicity island regulator protein interacts with several
trimeric dUTPases [2,3]. Upon dUTPase-Stl complex formation dUTPase enzymatic
activity is inhibited while the repressor function of Stl protein is also perturbed
[2,4]. It has been recently revealed that, in spite of its different structure
(Fig 1), dimeric dUTPase of NM1 phage also binds to Stl [5,6].
In this study, we attempted to answer how the interaction evolves between the
two highly divergent dUTPases and the Stl protein.
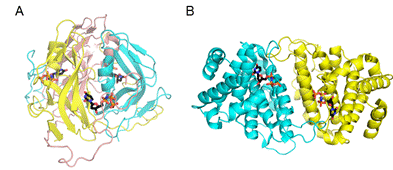
Figure 1. Comparison of the trimeric and dimeric dUTPase structures [3] A) Mycobacterium tuberculosis trimeric dUTPase B) Leishmania major dimeric dUTPase
We found that the dimeric dUTPase of the NM1 phage is inhibited by Stl with an apparent inhibitory constant of 34 ± 14 nM, which is comparable to that observed in case of trimeric dUTPases [7]. The stoichiometry of the NM1 phage dUTPase-Stl complex was investigated by native gel electrophoresis, chemical crosslinking and native mass spectrometry, both confirming that the interaction exists between Stl and NM1 phage dUTPase monomers. This indicates that in case of this dimeric dUTPase, Stl disrupts the oligomerization of the protein, contrary to the observations for trimeric dUTPases. These results can provide a possible explanation on the mechanism of enzymatic inhibition in case of the phage dimeric dUTPase, where the active centrum is at the dimer interface of the proteins (Fig 1), which is likely affected by Stl binding.
Based on the observed inhibition of the two markedly different dUTPases by Stl it was intriguing to hypothesize that this protein has a substrate mimicking surface which serves as a common interface for complexation with dUTPases. Consonant with this hypothesis it has been shown that the carboxi-terminal segment of Stl has a role in the interaction of the inhibitor protein with both the trimeric and dimeric phage dUTPases [6,8].
In order to investigate the specific
interaction surface of the proteins, we analyzed the change in hydrogen deuterium exchange (HDX) rate of those
upon complex formation with mass spectrometry (MS) [7].
The disruption of the oligomerization of the dimeric protein should be
considered during the interpretation of the HDX-MS results in case of the NM1
phage dUTPase. We found that the isotope exchange rate is close to constant in
the putative dimer interface, although it decreases in a segment adjacent to
that in the 3D model. Based on this we suggest that the dimer interface of NM1
phage dUTPase is covered by Stl and an additional segment of that is also
affected. We found that two different regions of the carboxi-terminal segment
of Stl show decrease in isotope exchange velocity in case of the dimer and
trimer phage dUTPase complexes. However since Stl also exists in a dimer
monomer equilibrium in solution, regions with unchanged exchange rate may also
contribute to the complex formation. As the structure of Stl either in the
dimer or in the monomer state is not yet determined, it is not yet possible to
locate these other segments probably involved in complex formation.
In conclusion, we found that two different regions of the Stl carboxi-terminal segment interact with the dimeric and trimeric phage dUTPases, although we cannot exclude that some other parts of the protein may additionally serve as a common interacting surface in different dUTPase-Stl complexes. Based on the differences we found we hypothesize that Stl interact with drastically different families of dUTPases in a different way, which is a hitherto unseen ability of an inhibitor protein.
Based on this knowledge it might be possible to design proteinaceous inhibitor(s) of other dimeric dUTPases essential for parasites as L. major or T. brucei causatives of leshmainiasis and African sleeping sickness, respectively.
1. K. Nyíri, B. G. Vértessy, BBA General Subjects, 1861, (2017), 3593.
2. J. E. Szabó, V. Németh, V. Papp-Kádár, K. Nyíri, I. Leveles, Á. Bendes, I. Zagyva, H. Pálinkás, B. Besztercei, O. Ozohanics, K. Vékely, K. Liliom, J. Tóth, B. G. Vértessy, Nucleic Acids Research, 42, (2014), 11912.
3. R. Hirmondó, J. E. Szabó,K. Nyíri, Sz. Tarjányi, P. Dobrotka, J. Tóth, B. G. Vértessy, DNA Repair 30, (2015), 21.
4. M. Á. Tormo-Más, I. Mir, A. Shrestha, S. M. Tallent, S. Campoy, Í. Lasa, J. Barbé, R. P. Novick, G. E. Christie, J. R. Penadés, Nature, 465, (2010), 779.
5. R. L. L. Hill, T. Dokland, J. Mol. Biol., 428, (2016), 142.
6. R. L. L. Hill, J. Vlach, L. K. Parker, G. E. Christie,
J. S. Saad, T. Dokland, J. Mol. Biol., (2017), in press
doi: 10.1016/j.jmb.2017.04.001
7. K. Nyíri, J. Matejka, M. J. Harris, A. J. Borysik, B. G. Vértessy, manuscript in preparation
8. K. Nyíri, B. Kőhegyi, A. Micsonai, J. Kardos, B. G. Vértessy, Plos One, 10, (2015), e0139086.
Authors acknowledge Dr. Terje Dokland for providing the plasmids encoding the gene of NM1 phage dUTPase.