Structure and function studies of RNA-binding proteins with FAST motifs and a RAP domain
J. Kutner, M. Merski, A. Jurska, M. Gorna
Structural Biology Group, Biological and Chemical Research Centre, Department of Chemistry, University of Warsaw, 101 Zwirki and Wigury St, 02-089 Warsaw, Poland
jkutner@chem.uw.edu.pl
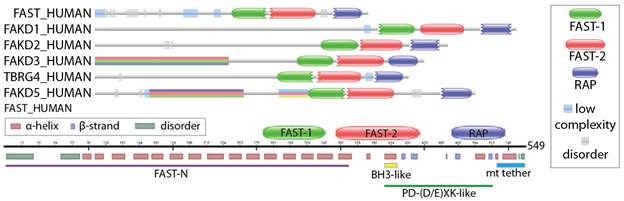
The FASTK family (Fas-activated Serine/Threonine Kinase) contains six human proteins which localize to the mitochondria and have been functionally linked to cellular respiration and with a rare mitochondrial disease. While human FASTK was initially annotated as an atypical Ser/Thr kinase later studies dispute this annotation [1]. Structurally, FASTKD proteins contain an N-terminal mitochondrial targeting signal, a pair of FAST motifs and a C-terminal RAP domain (Figure 1).The N-terminal part is predicted to be highly globular but with small disordered regions. The FAST motifs are putative RNA binding domains with a novel α-helical repeat fold that has no sequence similarity to any other known helical repeat motifs. Interestingly, the RAP domain is found in many members of the recently identified class of octotricopeptide repeat (OPR) proteins, which are abundant in plants and green algae and is believed to play a role in chloroplast RNA biology [2]. The OPR proteins have been shown to bind RNA with preference for some substrates [3], but their structure or RNA binding specificity is unknown. The RAP domain is also overrepresented in Plasmodium, and hence structural information of this domain is relevant to the field of malaria.
Figure 1. Domain composition and secondary structure prediction of the human FASTK family (Pfam: PF08373).
The obtained structures and functions of these proteins may have relevance to drug design therapeutic strategies, particularly of cancer and inflammation and will likely reveal new folds of RNA binding domains thus contributing to the general knowledge of the rules that govern RNA recognition. Our project aims to provide for the first time structural and novel biochemical information about the relatively understudied FASTK family.
1. C. Wu, M. H. Ma, K. R. Brown, M. Geisler, E. Tzeng, C. Y. Jia, I. Jurisica, S.S. Li, Proteomics, 7, (2007), 11.
2. A. H. Auchincloss, W. Zerges, K. Perron, J. Girard-Bascou, J. D. Rochaix, J Cell Biol., 157, (2002), 6.
3. M. Rahire, F. Laroche, L. Cerutti, J.D. Rochaix, Plant J. 72, (2012), 4.
Acknowledgements.
We acknowledge the financial support within the Polish NCN SONATA grant 2014/15/D/NZ1/00968.