Automated scoring of crystallisation experiments using multiple images
J. Wilson
Departments of Chemistry and Mathematics, University of York, York, UK
Finding the conditions that will produce diffraction quality crystals can require many crystallization experiments. The use of robots has increased the number of experiments performed in most laboratories and, in structural genomics centres, tens of thousands of experiments can be produced each day. Visual inspection is becoming increasingly impractical and automated imaging systems are now used routinely to record the results of these experiments. Image analysis software has been developed a number of research groups [1-4] to provide scores, allowing the images from crystallization trials to be examined in order of merit and reducing the number that need to be examined by eye. However, scoring individual images does not take advantage of the fact that each experiment is assessed regularly over a period of time. As each new image is produced, further information about the experiment becomes available and changes between images can be encoded as additional features for classification. The more information that can be obtained, the greater the likelihood of correct classification and, in addition to analysis of the time-course images as a sequence, the information gained from UV imaging is considered. For example, although the drop in figure 1 is easily identified in the greyscale gradient magnitudes, the drop in figure 2 cannot be found. However, the additional information from the UV image taken at the same time, allows a mask to be found so that further processing is restricted to the crystallisation drop.
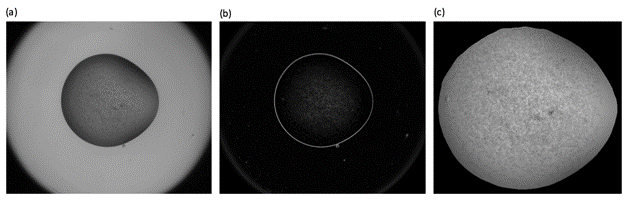
Figure 1. The greyscale gradient magnitudes (b) can be used to provide a mask for the crystallisation drop in (a).
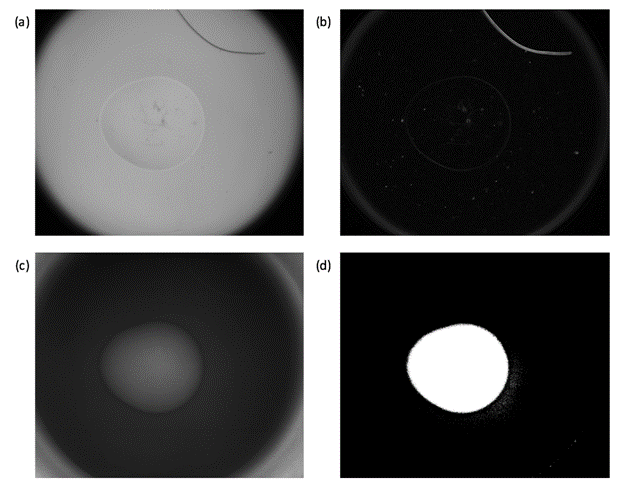
Figure 2. The lack of a defined boundary in (a) does not allow the drop to be identified by the gradient magnitudes (b). However, the UV image provides additional information that can be used to mask the drop.
1. S. Buchala, J. Wilson. Acta Cryst. D. Biol.Crystallogr., D64 (2008), pp. 823-833.
2. J.T. Ng, J.T., C. Dekker, M. Kroemer, M. Osborne, F.von Delft. Acta Cryst. D. Biol.Crystallogr., 70 (2014), pp. 2702-2718.
3. C. Cumbaa, I. Jurisica, I. J. Struct. Funct. Genomics, 11 (2010), pp. 61–69.
4. S. Pan, G. Shavit, M. Penas-Centeno, D.H. Xu, L. Shapiro, R. Ladner, E. Riskin, W. Hol, D. Meldrum. Acta Crystallogr. D. Biol. Crystallogr. 62 (2006), pp. 271–279.