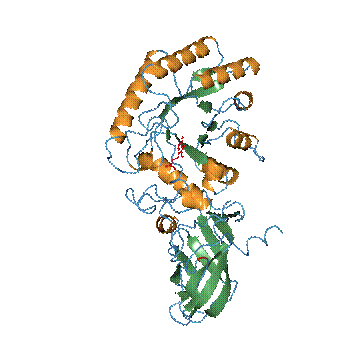Structural Comparison of Putative Alanine Racemases Alr and YlmE from Streptomyces coelicolor A3(2)
R. Tassoni1, L. van der Aart2, M. Ubbink1, G. P. van Wezel2, N. S. Pannu1*
1Leiden Institute of Chemistry, Leiden University, Gorlaeus Laboratories, Einsteinweg 55, 2333 CC Leiden, The Netherlands
2Institute of Biology Leiden, Leiden University, Sylviusweg 72, 2333 BE Leiden, The Netherlands
*raj@chem.leidenuniv.nl
The conversion of L-alanine into D-alanine in bacteria is essential for cell wall synthesis and bacterial survival, and performed by pyridoxal phosphate (PLP)-dependent enzymes called alanine racemases. Based on sequence annotation, the two genes alr and ylmE of Streptomyces coelicolor A3(2) are hypothesised to encode for proteins with alanine racemase activity. In the present study, we present the crystallization and crystal structures of both uncomplexed Alr and YlmE and in complex with ligands. The two proteins share a common a/b barrel fold, which characterizes the N-terminal domain of Alr and the whole YlmE peptide. In both proteins, the catalytic PLP-cofactor is bound to a lysine residue located in the core of the a/b barrel domain, Lys46 in Alr and Lys40 in YlmE. Despite these common features, YlmE lacks the C-terminal domain present in Alr and that mediates homodimerization by head-to-tail binding with the N-terminal domain. Furthermore, in vitro biochemical studies show alanine racemization only for Alr, with a rate of 2.12 µmol min-1 for the racemization of L- to D-Ala and 2.90 µmol min-1 for the opposite direction. Our results confirm the alanine racemase activity of Alr and open the way for further studies to elucidate the function of YlmE.
|
|
|
Figure 1. A ribbon representation of the Alr monomer. a-Helixes and b-strands are represented in orange and green, respectively. The PLP cofactor is shown as a ball-and-stick model in red. The catalytic Tyr283 is highlighted in red. |
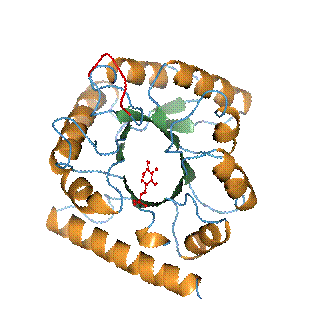
Figure 2. A ribbon representation of the YlmE fold. a-Helixes and b-strands are represented in orange and green, respectively. The PLP cofactor is shown as a ball-and-stick model in red. The residues highlighted in red correspond to a flexible loop region. |
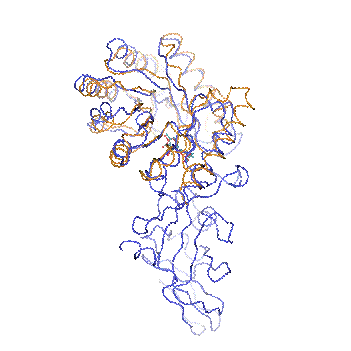
Figure 3. Superposition of
Alr (blue) and YlmE (orange). The PLP cofactors are shown as ball-and-stick
models and are coloured in dark cyan and red for Alr and YlmE, respectively.
YlmE superposes well with the N-terminal domain of Alr but lacks the C-terminal
domain responsible for dimerization and alanine racemization activity.