Crystallization of chromatin complexes
Song Tan, Robert McGinty, Makde Ravindra, Girish Taverekere, Ryan Henrici
Center for Eukaryotic Gene Regulation, Penn State University, University Park, PA 16802, USA
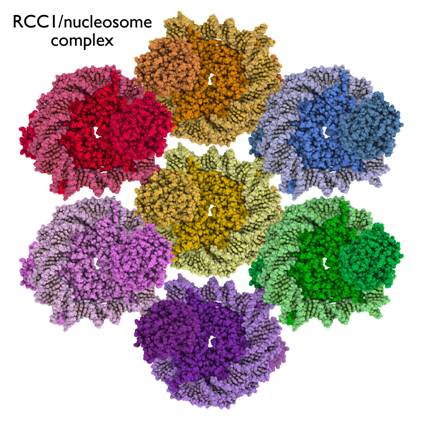
Chromatin complexes of chromatin factors or enzymes bound to the nucleosome offer multiple challenges common to crystallizing multicomponent macromolecular complexes: producing milligram quantities of homogeneous sample, growing crystals of the desired complex and improving diffraction of the crystals to high resolution. I will describe our efforts to crystallize different chromatin complexes, including the RCC1/nucleosome complex and the Polycomb PRC1/nucleosome complex, and what we have learned from these efforts. For the 300 kDa RCC1/nucleosome complex, varying the RCC1 (Regulator of Chromosome Condesation) species improved initial imperfect crystals to block single crystals which diffracted anisotropically and finally to block single crystals which diffracted isotropically. Post-crystallization soaks of crystals in dehydrating solutions made it possible to collect diffraction data to 2.9 Å. I will also describe procedures we use to optimize complex reconstitutions and to evaluate whether initial crystals contain the chromatin factor or enzyme.