X-ray Crystallography Studies of Protein Complexes Controlling Cell Cycle Gene Expression
P. Ramanan, K.Z. Guiley, A.H. Marceau, and S.M. Rubin
Department of Chemistry and Biochemistry, University of
California, Santa Cruz, CA 95064
srubin@ucsc.edu
Cell cycle gene expression is mediated by two transcription factor complexes known as DREAM and Myb-MuvB (MMB). DREAM represses cell cycle genes in quiescence and G1. Upon cell cycle entry, DREAM dissociates and MMB binds promoters to activate gene expression. While studies have identified the broad transcriptional roles of DREAM and MMB, the structure and biochemical function of these complexes have been poorly characterized. Both complexes share a core scaffold of five proteins known as MuvB, which binds DNA and histones. We are using x-ray crystallography to study the structure and function of MuvB and to understand how the association of MuvB with repressing and activating transcription factors is regulated. Our strategy relies on co-expression of multicomponent complexes and subcomplexes using the baculovirus system. We have successfully designed subcomplexes suitable for crystallization using structure prediction, co-precipitation, and limited proteolysis. X-ray structures of these subcomplexes have revealed how MuvB interacts with DNA, how DREAM and MMB are assembled, and how the complexes bind histones. Our structures and complimentary data support a novel nucleosome-positioning model for MuvB function.
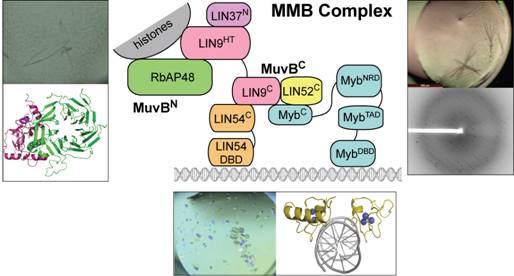
Figure 1. X-ray crystallography studies of the MMB complex. Schematic for the proposed domain organization is in the center. Crystals, diffraction data, and structures are shown for several subcomplexes.