In-solution structure and oligomerization of human histone deacetylase 6 - an integrative approach
Shivam Shukla1,2, Jan Komarek1, Zora Novakova1, Jana Nedvedova1, Kseniya Ustinova1, Pavla Vankova1, Alan Kadek3,4, Charlotte Uetrecht3,4,5, Haydyn Mertens6, Cyril Barinka1*
1Institute of Biotechnology of the Czech Academy of Sciences, BIOCEV, Prumyslova 595, Vestec, Czech Republic.
2Department of Physical Chemistry, Faculty of Natural Science, Charles University, Albertov 6, Prague, Czech Republic.
3Leibniz Institute for Virology (LIV), Martinistrasse 52, 20251 Hamburg, Germany.
4European XFEL GmbH, 22869 Schenefeld, Germany.
5Centre for Structural Systems Biology, Deutsches Elektronen-Synchrotron (DESY), Notkestrasse 85, 22607 Hamburg, and Department of Health Sciences and Biomedicine, School of Life Sciences, University of Siegen, Am Eichenhang 50, 57076 Siegen, Germany.
6European Molecular Biology Laboratory (EMBL)-Hamburg Outstation, c/o DESY, Notkestrasse 85, 22603 Hamburg, Germany.
e-mail: cyril.barinka@ibt.cas.cz
Histone deacetylases (HADCs) belong to the family of enzymes that remove the acetyl group from lysine side chains of target proteins regulating thus a plethora of cellular process. Among all other HDACs, HDAC6 is a large (140 kDa) and structurally complex multidomain enzyme harbouring a mosaic of unstructured and globular domains (Fig 1). Its primarily found in cytoplasm and acts on many non-histone targets including tubulin, Hsp90, and peroxiredoxins [1-5]. Structural data available currently are only on isolated globular domains and given its structural complexity, the full-length human HDAC6 is a challenging target for X-ray crystallography. To glean structural information on full-length human HDAC6, we used an integrative approach by combining experimental data from several orthogonal biophysical techniques including analytical ultracentrifugation (AUC), size-exclusion chromatography-multiangle light scattering (SEC-MALS), native mass spectrometry (MS), H/D exchange and small-angle X-ray scattering (SAXS). Our in-solution structural model shows that HDAC6 exists as an ensemble of conformers in solution. Furthermore, our data shed light on HDAC6 concentration-dependent oligomerization mediated by mannerist N-terminal domain. Overall, our findings can be used for further research into structure-function and physiological studies of this unique deacetylase
1. Zou, H., et al., Characterization of the two catalytic domains in histone deacetylase 6. Biochem Biophys Res Commun, 2006. 341(1): p. 45-50.
2. Skultetyova, L., et al., Human histone deacetylase 6 shows strong preference for tubulin dimers over assembled microtubules. Sci Rep, 2017. 7(1): p. 11547.
3. Hubbert, C., et al., HDAC6 is a microtubule-associated deacetylase. Nature, 2002. 417(6887): p. 455-458.
4. Kovacs, J.J., et al., HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell, 2005. 18(5): p. 601-7.
5. Bali, P., et al., Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem, 2005. 280(29): p. 26729-34.
6. Miyake, Y., et al., Structural insights into HDAC6 tubulin deacetylation and its selective inhibition. Nat Chem Biol, 2016. 12(9): p. 748-54.
7. Hai, Y. and D.W. Christianson, Histone deacetylase 6 structure and molecular basis of catalysis and inhibition. Nat Chem Biol, 2016. 12(9): p. 741-7.
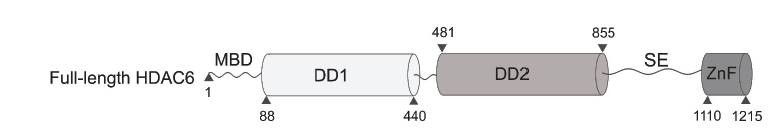
Fig 1: A schematic representation describing the domain organization of full-length HDAC6.
We acknowledge the Grant agency of the Charles University (Project No: 1678218) for their support.