Structural analysis of S1-like nuclease from opportunistic biofilm dwelling pathogen Stenotrophomonas maltophilia
K. Adámková1, 2, T. Kovaž1, M. Trundová1, B. Husáková1,2, J. Dohnálek1
1Institute of Biotechnology of the Czech Academy of Sciences, v.v.i., Průmyslová 595, 252 50 Vestec, Czech Republic
2University of Chemistry and Technology Prague, Department of Biochemistry and Microbiology, Technická 5, 166 28 Prague 6, Czech Republic
adamkovak@ibt.cas.cz
S1-P1-like nucleases are a family of zinc-dependent enzymes cleaving phosphodiester bonds of nucleic acids. Members of this family from fungi and plants have already been studied and are widely used in biochemistry and biotechnology [1]. However, members from pathogenic organisms, such as some bacteria and protozoan parasites, have not yet been characterized and their function is not fully understood. Knowledge of their structure, active site composition, substrate preferences, and cleavage mechanism could be an important step towards exploiting their biotechnology potential.
The subject of our study is a zinc-dependent nuclease from Stenotrophomonas maltophilia (SmNuc1), small globular protein with high activity against single-stranded DNA, double-stranded DNA, as well as RNA. Bacterium Stenotrophomonas maltophilia is an emerging multi-drug-resistant Gram-negative aerobe causing severe nosocomial respiratory infections in humans, primary infecting immunocompromised patients. These infections are often complicated by the ability of this opportunistic pathogen to form highly resistant biofilm on various surfaces [2].
Here we present a novel structure of recombinant SmNuc1 nuclease obtained at 1.4 Å resolution, followed by structures of complexes with DNA and RNA cleavage products, and structures of SmNuc1 mutants and their complexes with 5′-mononucleotides. Analysis of these high-resolution crystal structures (1.2 2.0 Å) combined with activity studies has expanded our knowledge of the active site composition and the impact of individual residues on the activity and substrate preferences. This study also revealed several interesting features, such as a flexible loop near the active site (we termed this loop R-loop because of the presence of the active‑site‑forming Arg74) capable of significant remodeling of substrate binding site, which brings up new questions about the catalytic mechanism. This information could shed light not only on some aspects of the SmNuc1 behavior, but also help us better understand the entire S1-P1 nuclease family.
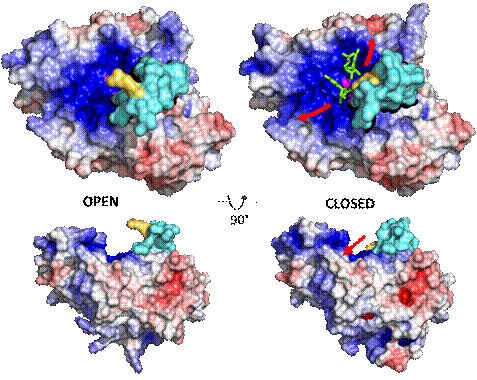
Figure 1: SmNuc1 solvent accessible surface. The cyan part of the surface indicates R‑loop near the substrate binding site, with Arg74 shown in yellow, and the rest of the surface is coloured by electrostatic potential (−5 kT/e 5 kT/e). Open R-loop is shown on the left and closed R-loop on the right. Red arrows indicate the direction of nucleic acid binding, ligands are shown in green sticks and zinc ions as magenta spheres. All graphics was created using PyMOL (Schrödinger).
1. Kovaž T, Dohnálek J, Biotechnology Advances, 2018, 36(3): 603-612
2. Brooke J. S., Clinical Microbiology Reviews, 2012, 25(1): 2-41
The work was supported by the institutional support of IBT CAS, v.v.i. (RVO: 86652036), ERDF (CZ.02.1.01/0.0/0.0/15_003/0000447 and CZ.02.1.01/0.0/0.0/16_013/0001776), MEYS CR (LM2018127, support of Biocev-CMS core facilities Crystallization of Proteins and Nucleic Acids, and Structural Mass Spectrometry of CIISB, part of Instruct-ERIC) and by specific university research (grant No A1_FPBT_2022_001).