REPLICATE, DIVIDE, MUTATE, SURVIVE: EVOLVING CRYSTAL STRUCTURES FROM PXRD
M. Tremayne1, S.Y. Chong1, C.C. Seaton2
1School of Chemistry, University of Birmingham, Edgbaston, Birmingham B15 2TT, UK
2School of Pharmacy, University of Bradford, Bradford, BD7 1DP, UK
The crystal structure determination of molecular materials from powder diffraction data has been driven in recent years by the development of direct space methods of structure solution [1]. These approach structure solution by generation of trial crystal structures, often based on the known connectivity of the material, and assessment of the fitness of each structure by comparison with the experimental data. Global optimization methods are then used to locate the global minimum corresponding to the structure solution.
In this paper we present the implementation of a global optimization technique based on the differential evolution algorithm (DE) [2]. DE is an evolutionary process that maintains a population of structures (or members) that are mutated and recombined together over a number of generations until the population converges on the global minimum. The combination of mutation and recombination in a single step to create a child (eqn. 1), and the deterministic method of selection by comparison of a child with its parent, means that DE is both relatively simple and easy to implement, while offering robust searching of minima. It can be controlled using only three optimization parameters (ie. K and F – the levels of recombination and mutation respectively, and the population size Np) and has been used in the solution of structures of intermediate complexity without the need for multiple runs, and in some cases required the generation of less than a thousand trial structures.
Child = Parent + K(Random1 – Parent) + F(Random2 – Random3) (1)
Application of the DE method will be illustrated by the structure solution of a range of organic materials including new polymorphic forms [3], families of compounds in which the primary interest centres on intermolecular aggregation [4] and supramolecular materials (Figure 1). All examples have been solved from powder diffraction data collected on a conventional laboratory diffractometer, often significantly affected by preferred orientation. The limitations of direct-space methods when faced with such detrimental sample characteristics will also be discussed.
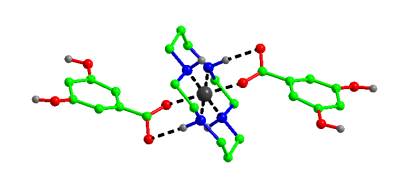
Figure 1: The crystal structure of Ni(cyclam)dihydroxybenzoate solved from PXRD by the DE method.
[1] K.D.M.Harris, M.Tremayne & B.M.Kariuki (2001). Angew. Chem., Int. Ed., 40, 1026.
[2] K.V.Price (1999). New Ideas in Optimization, D.Corne, M.Dorigo, F.Glover (Eds), McGraw-Hill, London UK, 77.
[3] C.C.Seaton & M.Tremayne (2002). Chem. Commun., 880.
[4] M.Tremayne, C.C.Seaton & C.Glidewell (2002). Acta Cryst. B58, 823.