CONFORMATIONAL BEHAVIOUR OF 11-O-METHYLAZITHROMYCIN IN THE SOLID AND SOLUTION STATE
Nada Koštia-Hulita,1 Dijana Matak-Vinkovic,1 Mladen Vinkovic,2 Gorjana Lazarevski2 and Gabrijela Kobrehel2
1 Faculty of Science, Univ. of Zagreb,
Zvonimirova 8, HR-10000 Zagreb, Croatia
2 PLIVA Research Institute, Baruna Filipoviaa 25,
HR-10000 Zagreb, Croatia
E-mail: dijana@zoak.pmf.hr
Keywords: antibiotic, macrolide, azalide,
azithromycin, 11-O-methylazithromycin, molecular
replacement, conformation, molecular mechanics
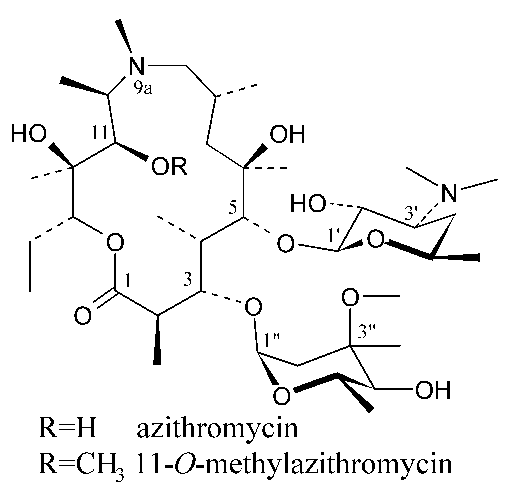
Azithromycin is the first, and medically the most important member of azalide class of macrolide antibiotics (1, 2). 11-O-Methylazithromycin is a new derivative of azithromycin with improved antibacterial activity (3). We present here the X-ray crystal structure analysis of 11-O-methylazithromycin DMSO solvate and comparison to its solution state conformation (4).
Unfortunately, single crystals of 11-O-methylazithromycin diffracted on Philips PW1100 four circle diffractometer only up to 1.3 A. Attempts to solve the structure by direct methods implemented in SIR97 and SHELXS97 program packages failed and we turned our attention to molecular replacement program AMoRe from CCP4 package. Search models were "folded-in" and "folded-out" solution state conformations of 11-O-methylazithromycin from our previous study (4). The highest solution from the rotation function yielded a peak 5.8 s above second highest peak for "folded-in" and 2.3 s for "folded-out" model. Therefore, "folded-in" model was further processed by translation function giving correct solution with correlation coefficient 69.2%. Structure was refined on F2 to RF=0.060 using SHELXL97 program and data up to 1.0 A. All non-H atoms were refined anisotropically, except for disordered solvent atoms.
This crystallographic analysis reveals that 11-O-methylazithromycin adopts "folded-in" conformation in the solid state. The same conformation dominates in CDCl3 solution (4). However, in the crystal 11-OMe group is oriented toward the centre of molecule ("11-OMe in" conformation) while in the CDCl3 solution it is placed in opposite direction, out of molecule ("11-OMe out" conformation). Molecular mechanics calculations suggest that "11-OMe in" conformation is more stable one, but appearance of higher energy "11-OMe out" conformer in solution state could be a consequence of specific intermolecular interactions with discrete CDCl3 molecules.
Crystal data: C39H74N2O12*C2H6OS,
FW=841.13, monoclinic, P21, Z=2, a=12.163(12),
b=8.879(9), c=23.17(2) A, b=91.79(5),
Dc=1.117 Mg m-3, l(MoKa)=0.71069 A, m=0.12
mm-1, RF=0.060 for 1264 data with Fo
> 4s(Fo), wRF2
=0.157 and S=0.973 for all 2835 data.