HYDROGENASES: HYDROGEN-METABOLIZING ENZYMES. WHAT DOES CRYSTALLOGRAPHY TELL US ABOUT THEIR MODE OF ACTION?
Juan Carlos Fontecilla-Camps1, Michel Frey1 , Elsa Garcin1, Claude Hatchikian2, Yaël Montet1, Claudine Piras1, Xavier Vernede1 & Anne Volbeda1
1Laboratoire de Cristallographie et de
Cristallogenese des Protéines, Institut de Biologie Structurale
Jean-Pierre Ebel CEA-CNRS, 41 Avenue des Martyrs 38027 Grenoble
CDX 1 France;
b Unité de Bioénergétique et
Ingéniérie des Protéines, CNRS, 31 Chemin Joseph Aiguier,
13402 Marseille CDX 20 France.
Keywords: hydrogenases/ hydrogen metabolism/ nickel enzyme/ biological electron and proton transfer/ iron-sulfur proteins
Hydrogenases are H2-activating metalloenzymes which are found in many bacteria. They have the unique ability to produce or to consume molecular hydrogen following the reaction:
H2 <=>2H+ + 2e- . A wealth of studies (EPR, Mossbaüer, Infrared, ENDOR, X-ray absorption, X-ray crystallography, organometallic chemistry, electrochemistry, molecular biology) on the structural and catalytic properties of the enzyme are currently pursued with in mind the design in the long range new strategies to produce cheaper hydrogen, using the protein as a model.
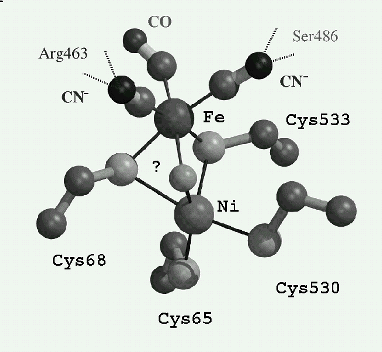
The determination in 1995, of the first three-dimensional crystal structure of an hydrogenase (at 2.85 A), [1] has given an enormous impetus to studies on this enzyme, including the determination of crystal structures of other hydrogenases and mutants. These have mainly led to the complete elucidation of the unusually complex structure of the active site, unprecedented in biology, since it consists of a two cysteines bridged Ni-Fe bimetallic site and three diatomic ligands to the iron, that is one (CO) and two (CN-); the two CN- being strongly hydrogen bonded to the protein [2,3; Figure]. Moreover, it has been shown that specific routes exist to transfer H2 [4], e- [1], and probably H+, between the molecular surface and the active site deeply buried in the protein .
It is also of interest to note that, in conjunction with crystallographic studies [2], it has been established that diatomic vibrational spectroscopy can report on electron density shifts occuring at the active site upon activation of the enzyme [3]. This provides a new and very powerful method to probe the catalytic activity of hydrogenases.

The active site of the oxidized hydrogenase from Desulfovibrio gigas (?) is putatively a m-oxo- species. Fe-Ni ~ 2.9 A
Due to the complexity of the active site, the catalytic mechanism is still highly debated. However the current models point to a mechanism where the nickel atom is most likely responsible for a base-assisted heterolytic cleavage of the hydrogen molecule whereas the iron atom is redox active.